James D. Moody,PhD

C206 BNSN, 801-422-6272
Email: james.moody@byu.edu
Protein engineering and structural biology to accelerate disease treatment
Like you and the entire biomedical enterprise, we want to help cure all of the diseases that plague Heavenly Father’s children! We use protein modeling, protein engineering, protein biochemistry, and protein structural biology to tackle bottlenecks in the drug and biologic development enterprise.
New tools to visualize protein structures at atomic resolution
If you’re trying to engineer a new drug or biologic, it’s best to start from an atomic level image of the protein the drug or biologic is intended to interact with. Once you have a drug or biologic candidate, it’s essential to see an atomic level image of that drug or biologic interacting with its target protein. Cryo-electron microscopy is a powerful new technique, but it’s not good for getting high resolution images of proteins smaller than 60–100 Å in size. X-ray crystallography is perfect for getting high resolution images of these smaller proteins. The only problem is that for X-ray crystallography to work, you have to get your protein of interest to form a well-ordered crystal, which has about a 10% success rate!
Protein of interest —> Crystals —> X-ray Diffraction data —> Electron Density —> Protein structure

Protein of interest

Crystals

X-ray Diffraction data
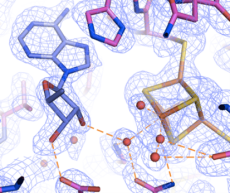
Electron Density
For the past several years we’ve been investigating and improving TELSAM fusion crystallization, a new tool to make disease proteins crystallize so we can get those precious atomic level images. TELSAM, engineered from a normal human protein, is soluble at pH >8 but forms a helical polymer at pH <7. You simply genetically fuse TELSAM to your protein of interest, purify the fusion protein, and lower the pH to polymerize TELSAM. The TELSAM polymers (with your protein of interest in tow) readily form nice crystals, with a >90% success rate!


Top view of TELSAM--target protein crystal lattice

side view of TELSAM--target protein crystal lattice
What else do we do?
We also do Cryo-electron microscopy of large, never before seen protein machines, we develop novel biologics, and we develop novel small molecule drugs
Interested in joining?
In the Moody lab you can learn computational protein modeling and design, molecular biology techniques, protein biochemistry, protein X-ray crystallography and protein cryo-electron microscopy. If you’re interested, I’d love to talk with you! We welcome dedicated, hardworking students with all levels of experience, Freshman through Senior. You should be able to spend at least 10 hours per week during Fall and Winter semesters in the lab to make meaningful progress.
References
1. Nawarathnage S, Soleimani S, Mathis MH, Bezzant BD, Ramírez DT, Gajjar P, Bunn DR, Stewart C, Smith T, Pedroza Romo MJ, Brown S, Doukov T, Moody JD. Crystals of TELSAM-target protein fusions that exhibit minimal crystal contacts and lack direct inter-TELSAM contacts. Open Biology. 2022 Mar;12(3):210271.
2. Gajjar PL, Pedroza Romo MJ, Litchfield CM, Callahan M, Redd N, Nawarathnage S, Soleimani S, Averett J, Wilson E, Lewis A, Stewart C, Tseng YJ, Doukov T, Lebedev A, Moody JD. Increasing the bulk of the 1TEL-target linker and retaining the 10×His tag in a 1TEL-CMG2-vWa construct improves crystal order and diffraction limits. Acta Crystallogr D Struct Biol. 2023 Oct 1;79(Pt 10):925-943.
3. Nawarathnage S, Tseng YJ, Soleimani S, Smith T, Pedroza Romo MJ, Abiodun WO, Egbert CM, Madhusanka D, Bunn D, Woods B, Tsubaki E, Stewart C, Brown S, Doukov T, Andersen JL, Moody JD. Fusion crystallization reveals the behavior of both the 1TEL crystallization chaperone and the TNK1 UBA domain. Structure. 2023 Sep 25:S0969-2126(23)00326-X.